 Antibody production
Antibody production
Interview of Pr. Bérangère BIHAN-AVALLE, expert in Phage Display Technology
Bérangère BIHAN-AVALLE, researcher at the Enzyme and Cell Engineering Unit UMR 7025 CNRS (GEC) and expert in Phage Display Technology, speaks about her fascinating discoveries in catalytic antibodies.
- What is your academic background? What led you to your actual researches?
- Thanks to the Phage Display technology, you identified catalytic antibodies (abzymes), in particular with β‐lactamase activity, which are very different from traditional antibodies with binding activity only. How do you explain such achievement?
- Which strategy did you use?
- Have you tested these abzymes in vitro or in vivo? if so, what are the results and the models used?
- What is the next step? What are the applications of the discovered abzymes?
What is your academic background? What led you to your actual researches?
After graduating from Paris University Denis Diderot (Paris VII) with a degree in immunology, I received my PhD from the University of technology of Compiègne (UTC, France) where I generated catalytic antibodies using the idiotypic network. I then experienced a 3-years post-doctoral fellowship in UCLouvain (Belgium) with Pr Jacques Fastrez where I developed phage display selection processes based on catalytic efficiencies and activities. At that time, the lab was a pioneer in the field of phage display and selections of proteins based on their catalytic activity were innovative. Back to UTC to become a lecturer, I brought my expertise in the field of selections and designs of libraries of various types (aptamers, peptides and antibodies) used for various purposes like fundamental research on auto-immune diseases and drug discovery.
Thanks to the Phage Display technology, you identified catalytic antibodies (abzymes), in particular with β‐lactamase activity, which are very different from traditional antibodies with binding activity only. How do you explain such achievement?
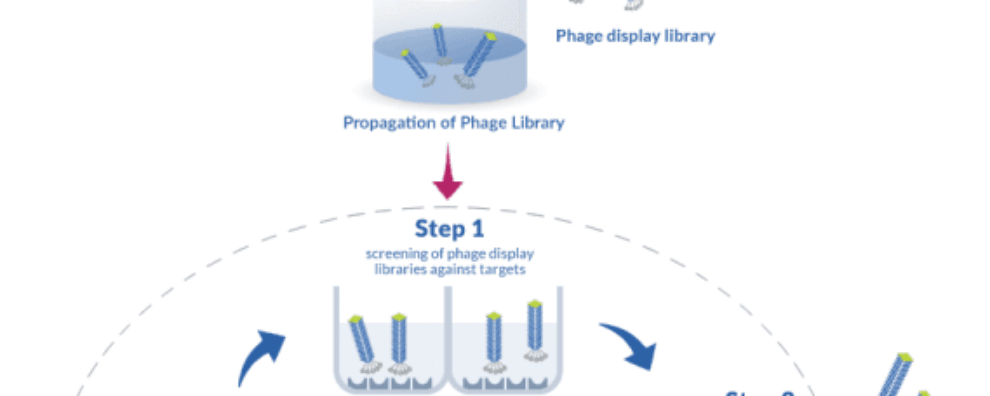
Catalytic antibodies are capable of catalyzing chemical transformations. The plasticity of antibodies and their ability not only to bind antigens but also to hydrolyse targets is exemplified by the generation of abzymes. Their involvement in diseases, especially in auto-immune ones, has been the subject of numerous studies and despite advances in the field, their origin and their physio-pathological role is still to be understood. That is why we conducted research on the capability of auto-immune prone mice to produce more catalytic antibodies than healthy ones. For that purpose, we used the technique of phage display to construct antibody libraries from the spleens of mice with healthy and autoimmune background, both with different immunological status: one group of mice was immunized with an antigen likely to induce catalytic antibodies, the other group remained naïve. We took b-lactamase as a model because, as well as proteases and peptidases, it hydrolyzed amide bonds. This activity is often found in auto-immune diseases. Phage display is of course a powerful tool allowing the in vitro reproduction of immune repertoires but the main challenge is to be able to select antibodies endowed with catalytic activity, not only binders.
Which strategy did you use?
To promote the generation of abzymes, we immunized a mice group with a homemade mechanism-based inhibitor of b-lactamase. This is called ‘reactive immunization’ because the goal is to trap catalytic antibodies during the maturation process of the adaptative immune response. We carefully designed primer sequences in order to amplify most of the generated variable regions, to get libraries of single-chain fragment variable (scFv) close in diversity to natural repertoires. Design of primer sequences and use of proper cloning strategy is the one of the critical step of the construction of antibody libraries on phages. Then we chose two targets for the selection procedure: one target was the mechanism-based inhibitor used for the immunization process, and the other one was a cyclic heptapeptide acting as a competitive inhibitor of b-lactamase. We proceeded by iterative selection, the target being immobilized on plates. This is where the watchful eye of an expert is important: experimental conditions have to be finely tuned to get what you search for.
Have you tested these abzymes in vitro or in vivo? if so, what are the results and the models used?
First, prior to biopanning, we have carried out a in silico study from the results of a high throughput sequencing of 400 000 scFv genes present in our libraries to highlight different expression patterns between healthy and auto-immune prone mice. We found a number of statistical differences that we have just published in Scientific Reports. Then, we tested the activity of the selected antibodies in vitro by using chromogenic and fluorogenic substrates but unfortunately, we were able to get only five catalytic antibodies, which is not statistically relevant.
What is the next step? What are the applications of the discovered abzymes?
We plan to repeat the Next Generation Sequencing using the latest technologies in order to have sequences covering the wholeness of the diversity of the libraries. We also wish to refine the experimental conditions of the selection procedure to get more than 5 abzymes as a result. We are also looking at achieving other objectives with other enzymatic activities such as proteases or DNases. In our research work, bioinformatics is becoming increasingly relevant, that is why we first speculate about the appropriateness of our approaches using in silico tools.
Our selected antibodies have no application but to clarify the link between auto-immune diseases and natural abzymes, but this knowledge will help to discover new treatments as for example in multiple sclerosis where the deleterious effect of catalytic antibodies was previously shown.