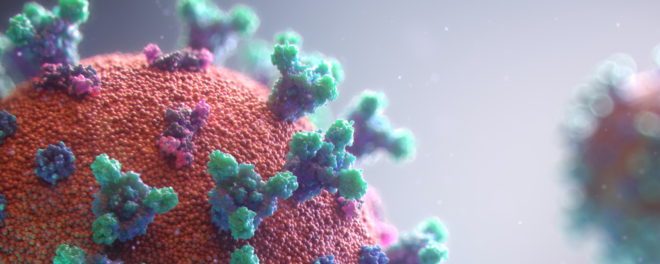
 Antibody production
Antibody production
Novel Coronavirus 2019 – How to Mitigate the Impact of the New and Future Epidemics?
News of the latest coronavirus outbreak continues to capture the world’s attention. Scientists have provisionally named it 2019-nCoV and, at the time of writing, this new strain has infected more than 4,000 people and killed more than 100 in the Hubei Province (China) alone. Although the country imposed a strict quarantine causing the lockdown of 56 million people, infections have already been reported in France, the United States, Canada, Australia, Thailand, and Nepal, among others.
The new coronavirus – 2019-nCoV
The symptoms associated with the new virus are not very different from those caused by common flu. But the morbidity and mortality rates have put the scientific and the global health care community in an uproar. Scientists have identified 2019-nCoV as a new member of the coronavirus family. A family with a track record for causing severe global-scale epidemics.
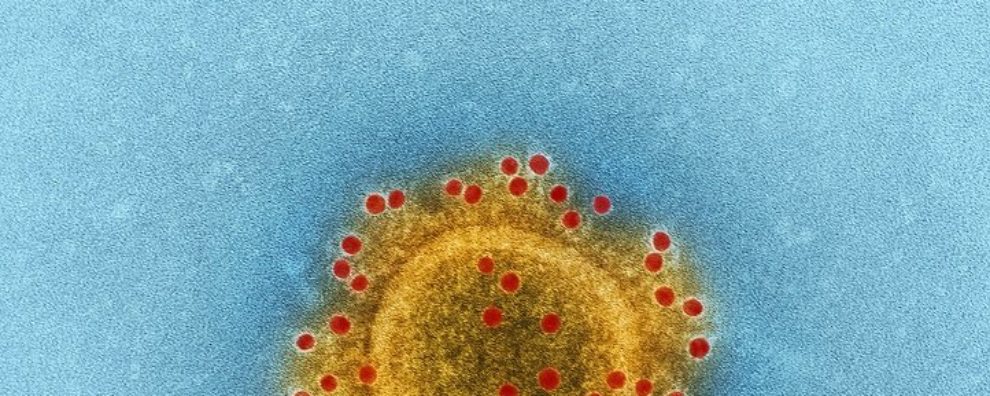
Coronavirus (CoV) belongs to the order Nidovirales, which is known to possess the largest and most complex RNA genomes ever described to date (up to 32 kb positive-sense RNA). Its RNA genome allows it to quickly adapt to new hosts from different species. Its name – CoV – originates from the virus’ morphology – crown-like spikes coating its surface.
The first human CoV was first identified in the mid-1960s, but it was only recently that its potential for causing outbreaks on a global scale has been established. The most devastating examples of CoV epidemics were the SARS (severe acute respiratory syndrome) and the MERS (Middle East respiratory syndrome) outbreaks, in 2002 and 2012, respectively.
Evidence suggests that both SARS and MERS virus originated from bats and presumed to have undergone extensive recombination with CoV from other animals before infecting humans. Like the new CoV, SARS and MERS can also spread from person-to-person through small or large aerosolized droplets originated from infected people’s coughs and sneezes.
Lessons learned from previous CoV epidemics
Despite the high mortality rates, CoV spreads like a common cold and afflicts more severely people with debilitated or suppressed immune systems. This was the case for SARS and MERS, which affected mostly people over 50 with diabetes, HIV infections, or kidney and liver problems. Although this tendency has yet to be confirmed for 2019-nCoV, there is still much to learn from SARS and MERS epidemics.
As of January 23, the Coalition for Epidemic Preparedness Innovations (CEPI) announced an investment of 12.5 million dollars to develop a vaccine aimed at containing the spread of 2019-nCoV. However, realistically even the most high-performing technologies won’t be able to put a new vaccine in the market in less than 1 year.
For this reason, vaccine development is a long-term measure to contain the new CoV. In the short term, the best strategy that can be used to mitigate the devastating effects of this disease is “implementing a holistic approach to diagnostics preparedness” (Kelly-Cirino, et al. 2019).
The conclusion comes from an extensive study led by Dr. Catharina Boehme from the Foundation for Innovative New Diagnostics (Geneva, Switzerland). The study reported the limited availability of proper diagnostics as one of the main catalysts behind the large-scale reach of these diseases.
This is one of the main differences between SARS and MERS, and the new CoV epidemics. As of January 25, 2020, at least 28 complete genomes and 1 partial genome of the new virus have already become publicly available through the NCBI (National Center for Biotechnology Information). This data allowed two things: the in silico design of effective diagnostic tools, and the study of the evolutionary history of the new virus.
The whole-genome comparison between these genomes allowed researchers to identify conserved regions in this diverse subpopulation. This led to the design of probes and primers that can specifically detect the new variant of human CoV.
Subsequently, the data prompted the development and validation of PCR-based assays for the quick and accurate diagnostics of the new disease. All suitable protocols developed by teams of experts around the world are available on the WHO website in its interim guide named – Laboratory testing for 2019 novel coronavirus (2019-nCoV) in suspected human cases.
The analysis of this data has also aided researchers in timing the first appearance of the new CoV. Although the conclusions are, to this date, provisional, scientists estimate the virus originated as early as November 2019. Surprisingly, this timing doesn’t seem to fit with the initial reports declaring the Wuhan seafood market as the source of the new disease.
Diagnostics technologies to contain the spread of devasting epidemics
Many different technologies can be used to detect and monitor the progression of infectious diseases. Generally, the most commonly used approaches include culture-dependent and culture-independent methods. The former is known to be an inexpensive but lengthy process. For this reason, it finds limited application in efforts to contain fast-progressing epidemics like human CoV.
On the contrary, culture-independent methods comprise a wide array of technologies that allow the quick detection of the pathogenic agent in suspected human cases.
Generally, these methods are based on PCR, ELISA or next-generation sequencing technologies. In the case of 2019-nCoV, researchers are focusing on real-time fluorescence-based PCR using either RNA directly extracted from potentially infected individuals or cDNA synthesized from these extracts. The easy access to data regarding 2019-nCoV has prompted the development of commercial kits for 2019-nCoV by many companies around the world.
Despite the inherent costs of PCR reagents, detection by this technology is likely to remain the favored method of detection of the 2019-nCoV in the upcoming months. The reason for this is that this technology can detect new infections in only a few hours and that specific primers and probes can be quickly synthesized in vitro.
However, real-time PCR is an extremely sensitive technology known to suffer from variability and reproducibility issues when different reagents and/or equipment are used. For this reason, it is likely that after the initial outbreak, researchers may focus on the development of more robust assays that can be performed by non-qualified personal.
Hence, ELISA assays for diagnostics may probably comprise the “second wave” of 2019-nCoV diagnostics. This “second wave” will be vital for controlling future outbreaks of the disease in case it becomes seasonal like the flu.
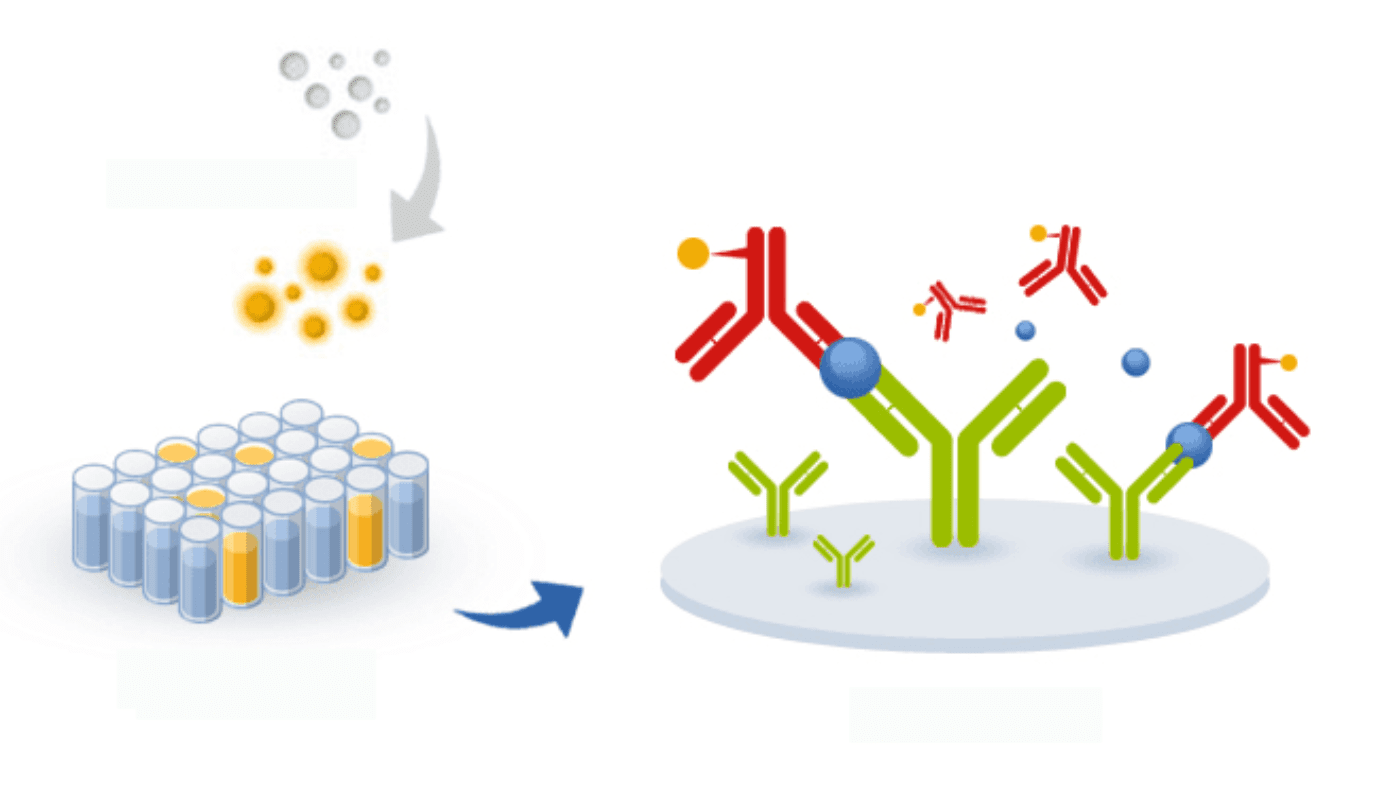
ELISA-based assays have been successfully developed for the inexpensive and user-friendly detection of SARS and MERS in potential patients. The same principles can be applied to the development of ELISA tests for the detection of 2019-nCoV. Unlike PCR, ELISA has the potential for parallelization and, thus, it can be adapted to allow the detection of multiple variants of the virus in a single plate and assay.
For this reason, although ELISA may be slightly less sensitive than PCR methods, if researchers succeed in creating an ELISA-based method for the detection of coronavirus, it may be possible to detect future outbreaks of the virus much earlier than has been possible for SARS, MERS, and even for the new 2019-nCoV.
- Andersen, K. Estimates of the clock and TMRCA for 2019-nCoV based on 27 genomes. Virological. Jan. 26, 2020. Available from: http://virological.org/t/clock-and-tmrca-based-on-27-genomes/347
- Boonham, N. et al. Methods in virus diagnostics: from ELISA to next generation sequencing. Virus Res. 2014; 186:20-31. doi: 10.1016/j.virusres.2013.12.007
- Chen, S. et al. Double-antigen sandwich ELISA for detection of antibodies to SARS-associated coronavirus in human serum. Eur J Clin Microbiol Infect Dis. 2005; 24(8):549-553. doi: 10.1007/s10096-005-1378-7
- Chen, Y. et al. A sensitive and specific antigen detection assay for Middle East respiratory syndrome coronavirus. Emerg Microbes Infect. 2015; 4(4): e26. doi: 10.1038/emi.2015.26
- Cohen, J. Scientists are moving at record speed to create new coronavirus vaccines—but they may come too late. Science. Jan. 27, 2020. Retrieved from: https://www.sciencemag.org/news/2020/01/scientists-are-moving-record-speed-create-new-coronavirus-vaccines-they-may-come-too
- Cohen, J. Wuhan seafood market may not be source of novel virus spreading globally. Science. Jan. 26, 2020. Retried from: https://www.sciencemag.org/news/2020/01/wuhan-seafood-market-may-not-be-source-novel-virus-spreading-globally
- Denison, M. R. et al. Coronaviruses – an RNA proofreading machine regulates replication fidelity and diversity. RNA Biol. 2011; 8(2): 270–279. doi: 10.4161/rna.8.2.15013
- Ellerin, T. The new coronavirus: What we do — and don’t — know. Harvard Health Blog. Jan. 25, 2020. Retrieved from: https://www.health.harvard.edu/blog/the-new-coronavirus-what-we-do-and-dont-know-2020012518747
- Kelly-Cirino, C. D. et al. Importance of diagnostics in epidemic and pandemic preparedness. BMJ Global Health. 2019; 4:e001179. doi: 10.1136/bmjgh-2018-001179
- Quammen, D. We Made the Coronavirus Epidemic. The New York Times. Jan. 28, 2020. Available from: https://www.nytimes.com/2020/01/28/opinion/coronavirus-china.html
- WHO Team. Laboratory testing for 2019 novel coronavirus (2019-nCoV) in suspected human cases. Interim guidance. WHO. Jan. 17, 2020. Retrived from: https://www.who.int/health-topics/coronavirus/laboratory-diagnostics-for-novel-coronavirus