 Antibody production
Antibody production
Why phage display for monoclonal antibody production remains an unrivaled technology
Phage display technology revolutionized antibody discovery. By coupling the structural simplicity of phages with the fast replication and high turnover of bacterial systems, the technology has shortened antibody development timelines well below historical levels. Currently, phage display for monoclonal antibody production enables the fast screening of large repertoires from naïve or immune hosts obtained by amplifying antibodies-genes expressed by peripheral blood cells. What challenges will shape the future of this technology? Check our frequently asked questions (FAQs) page about phage display for a complete overview of all steps of this robust process for antibody generation.
How phage display changed monoclonal antibody production?
In vitro phage display was first described by George Smith in the 1980s using filamentous phage M13. With the purpose of studying peptide-peptide interactions, fragments were fused to the phage’s capsid protein and displayed on its surface. This simple system allowed Smith and his team to cost-effectively select specific peptides within large libraries with the highest affinity towards a given target.
Later, Smith and his colleague Stephen Parmley capitalized on the phage’s simplicity by coupling peptide selection with multiple rounds of peptide enrichment. The process involved exposing phage-displayed peptides to immobilized targets, washing low-affinity binders, and infecting bacteria (Escherichia coli) with the remaining phages. When repeated multiple times, the cycle favors the propagation of phages fused to high-affinity peptides while gradually eliminating low-quality and low-affinity molecules.
These cycles became known as biopanning, and they allow screening large repertoires by leveraging the high propagation rate of M13 in E. coli. The technology was further improved and adapted to antibody display by researchers at The Scripps Institute in the early 1990s. Today, phage display for monoclonal antibody production is one of the few technologies that allows the generation of fully antibodies without relying on animal immunization.
Key principles of phage display for monoclonal antibody production
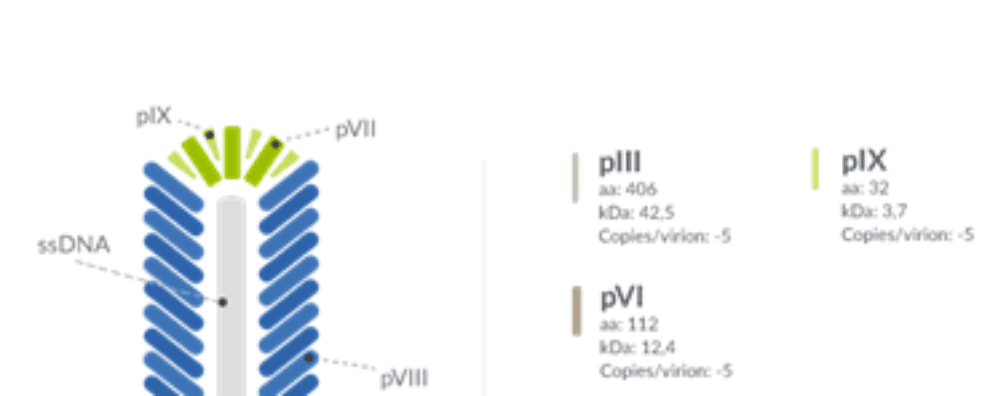
Although many systems have been tested, E. coli’s filamentous phage M13 continues to be the preferred phage system for antibody discovery. M13 belongs to a group of filamentous phages with a restricted infection range, infecting only E. coli strains expressing the F pilus consisting of a tubular structure with a key role in bacterial conjugation.
M13 contains a circular single-stranded DNA genome (ssDNA) and, unlike other phages, it displays neither a temperate nor a lytic behavior, instead, causing a chronic infection in its host. This infection works by triggering the continuous synthesis and release of new particles without compromising the integrity of its host, an invaluable property for performing multiple enrichment cycles (i.e. biopanning).
Typically, the process of phage display for monoclonal antibody production starts by amplifying antibody-encoding genes and ligating the corresponding PCR products to a phage display vector. Both for peptide interaction studies and antibody discovery, foreign molecules are typically fused to the major coat protein (gene pVIII) or the minor coat protein (gene pIII).
The success of this process largely depends on the quality and diversity of the antibody repertoire, which, in turn depends on the quality and source of the antibody information. For antibody discovery, this information is typically collected by isolating RNA from B cells or peripheral blood mononuclear cells. This is subsequently transcribed into cDNA which, in turn, serves as the template for amplification using a defined set of primers aimed at capturing the entire diversity of the host.
The library can be constructed to include different antibody isotypes (e.g. IgG, IgA, and IgE) and in different antibody formats: Fab (antigen-binding fragment) and scFv (single-chain antibody fragments). The latter is the most popular format for phage display, and it contains the heavy and light chains of the variable region (VH and VL) joined by a linker.
The main advantages of phage display in comparison to in vivo antibody generation entail the short timelines associated with this technology and possibility to use toxic and/or non-immunogenic antigens for antibody generation.
Improving methodologies of phage display for monoclonal antibody production
Overall, in vitro display of antibodies still faces two important challenges:
- The lack of natural affinity maturation – in naïve antibody repertoires (no immunization) this may on occasion translate into a weaker affinity towards a specific antigen than hybridoma-derived antibodies
- Loss of natural pairing information – in both naïve and immune libraries, heavy and light chains are paired randomly which, on occasion, may result in lower stability in comparison to hybridoma-derived antibodies
These reflect the current technical constraints in reproducing antibody affinity maturation in vitro. Currently, these challenges are successfully bypassed by using phage display to further increase the affinity of in vitro-derived antibodies (i.e. phage display of random or targeted mutate libraries), but can this current approach be further improved?
Like other techniques, phage display for monoclonal antibody production has considerably improved since its discovery in the 1990s. In recent years, one way to overcome these limitations has been to improve library quality by dramatically increasing the diversity, improving library construction methods, and including early in silico evaluation of promising binders.
Soon, it may be possible to further decrease current production costs and reduces development timelines, while fully leveraging the fast screening capacity of phage display for monoclonal antibody production.
Concluding remarks
Phage display technology is a robust approach to monoclonal antibody production. Reported in the 1990s, the technology quickly became one of the dominant technologies used for the discovery of fully human therapeutic antibodies.
Few techniques have the unique advantages of phage display. With improved library design, phage display technology is predicted to be increasingly important for therapeutic and analytical antibody discovery. To this day, its robustness, screening capacity, and tolerance towards toxic and non-immunogenic antigens continue to be unrivaled by other in vitro or in vivo techniques.
- Hammers, C. M. and Stanley, J. R. Antibody Phage Display: Technique and Applications. J Invest Dermatol. 2014; 134(2): e17. doi: 10.1038/jid.2013.521
- McCafferty, J. et al. Phage Antibodies: Filamentous Phage Displaying Antibody Variable Domains. Nature. 1990; 348(6301):552-554. doi: 10.1038/348552a0
- Rickert, K. W. et al. Combining phage display with de novo protein sequencing for reverse engineering of monoclonal antibodies. MAbs. 2016; 8(3):501-512. doi: 10.1080/19420862.2016.1145865
- Tiller, T. et al. A Fully Synthetic Human Fab Antibody Library Based on Fixed VH/VL Framework Pairings With Favorable Biophysical Properties. MAbs. 2013; 5(3):445-470. doi: 10.4161/mabs.24218
- Winter, G. et al. Making Antibodies by Phage Display Technology. Annu Rev Immunol. 1994; 12:433-455. doi: 10.1146/annurev.iy.12.040194.002245