 Antibody production
Antibody production
How nature inspires the development of better antibody design techniques
Antibody design techniques are recurrently used to improve or fine-tune the affinity, specificity and stability of these biomolecules. Most of these techniques draw inspiration from natural protein interactions and enzymes. However, the modifications to the antibody backbone can often have unforeseen effects on its effectiveness and specificity.
Key properties to consider during antibody design and optimization
Antibodies are complex molecules with countless therapeutic applications.
They can be generated in vivo (i.e. hybridoma technology) or in vitro (i.e. phage display technology) but their properties often need and should be further optimized after this initial stage.
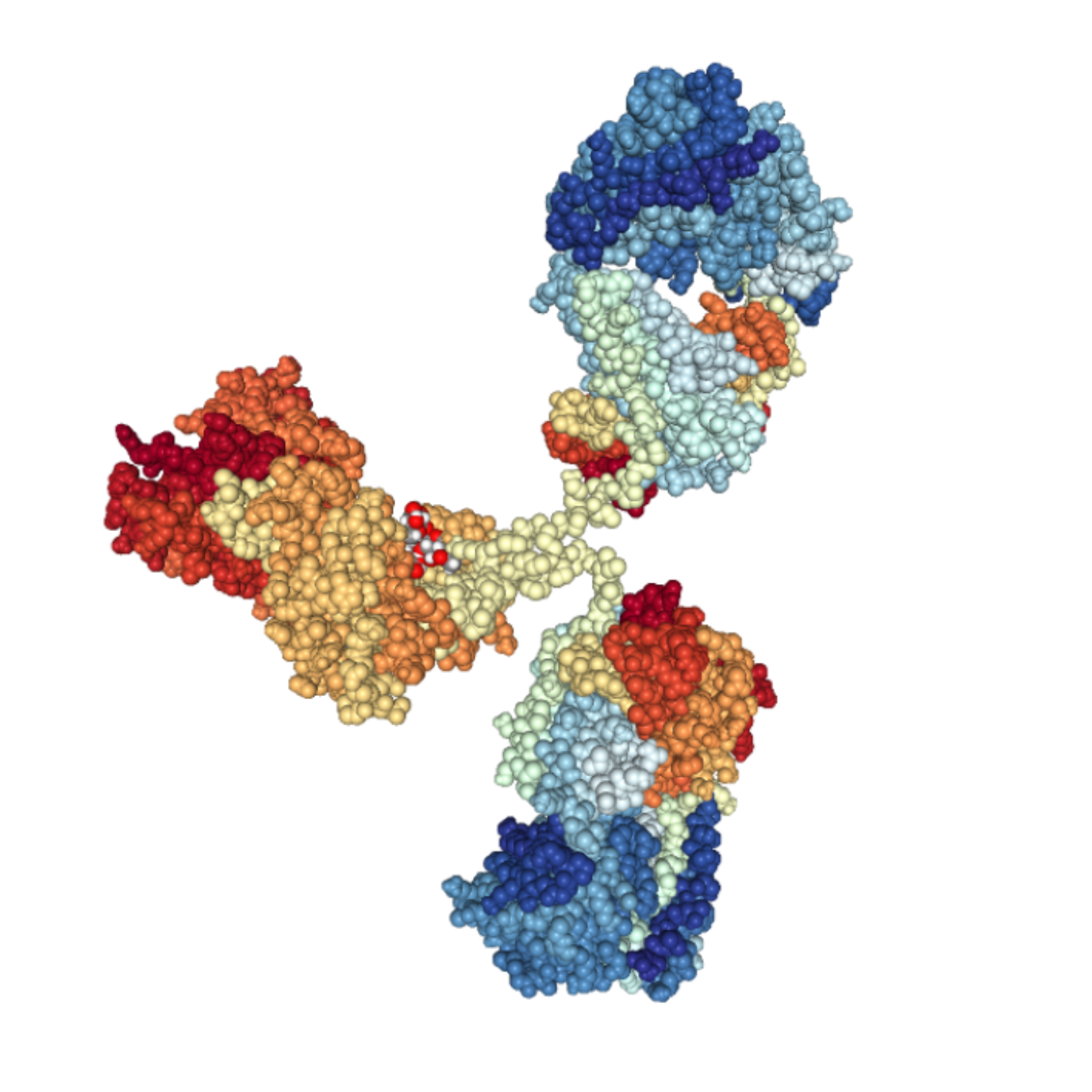
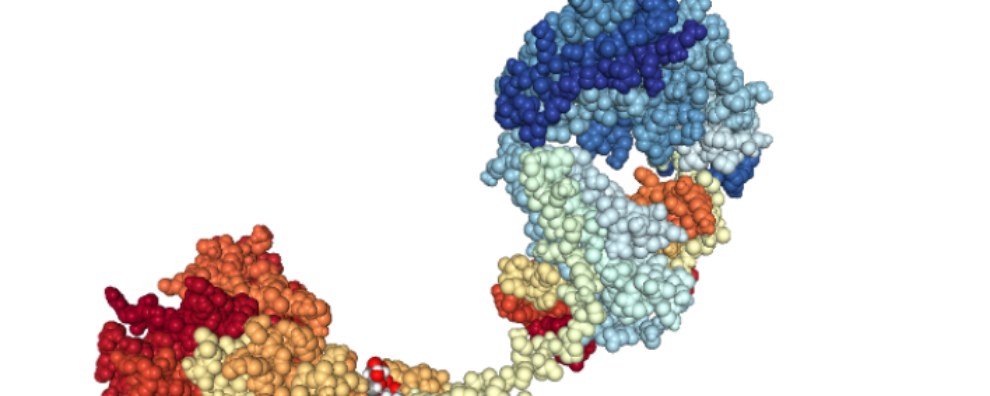
Nowadays, it is already possible to fine-tune several key properties of newly generated antibodies. For instance, researchers can now optimize the binding affinity, specificity, stability, solubility, pharmacokinetics, and effector functions of monoclonal and bispecific antibodies.
Moreover, specific antibody configurations such as antibody-drug conjugates can be optimized by addressing the potential issues raised by the use of chemical linkers to bind antibodies to specific drugs.
Nevertheless, regardless of the antibody format, the most important properties to be considered during antibody design are an antibody’s affinity and specificity.
Today, several approaches are currently and recurrently applied to the optimization of these properties:
- De novo design
- Design by mimicking natural protein interactions
- Directed evolution
- A combination of all of the above
Successful antibody design strategies to optimize affinity and specificity
De novo prediction is considered the most challenging approach to antibody design.
Several researchers have tested the optimization of the stability of antigen-antibody complexes using solely computational methods. However, to date, there is little experimental evidence to support the robustness of these methods.
For this reason, many researchers prefer to draw inspiration from natural protein interactions or to combine antibody design with directed evolution methods.
The first of these approaches can be made by seeking naturally occurring self-aggregation-prone proteins. This propriety makes these proteins suitable candidates for the grafting and combination of peptide segments into the antibody backbone. The rationale behind this strategy is the opportunity to take advantage of this aggregation-prone behavior to increase an antibody’s affinity towards a self-aggregating target.
Many of these aggregation-prone proteins are commonly linked to neurological debilitating diseases such as Alzheimer’s and Parkinson’s diseases.
Thus, some researchers have tried leveraging the pathogenic protein aggregation for the design of β-amyloid targeting antibodies.
An example of a recent success using this approach was the design of several antibodies using peptide regions of the Alzheimer’s Aβ42 peptide.
These antibodies, also named gammabodies (Grafted AMyloid-Motif antibodies), were designed to contain specific hydrophobic segments of the Aβ protein in their complementary determining region 3 (CDR3). Some of these constructs showed improved affinity towards the Aβ protein, while others did not.
These results showed that even when researchers draw their inspiration from natural interactions, it may be hard to predict which peptide segments will improve the overall affinity and specificity of an antibody.
Thus, many researchers focus on using directed evolution methods and combine them with semi-rational design.
These hybrid approaches consist in designing some residues according to computational predictions while randomizing others. This approach creates an antibody library that can quickly be screened using display techniques, such as phage display.
Moreover, this hybrid approach also provides the opportunity to insert residues able to recognize chemical modifications in antigens. Many of these post-translational modifications, such as phosphorylation, are associated with pathological conditions making them suitable targets for the design of new immunotherapies.
In a study, researchers from the Babraham Institute (Cambridge, UK), attempted to introduce phosphate-binding motifs from kinases into the CDRs of antibodies. They combined this approach with further randomized evolution and they screened the antibody library with phage display.
This approach allowed them to obtain several antibodies with improved affinities (40-5,000 nM) towards modified serine, threonine and tyrosine residues. Furthermore, it demonstrated the potential of these hybrid approaches for the design of better antibodies.
Advances in computational approach to antibody design
Despite the slow progress in the development of solid and reliable computational strategies for antibody design, many exciting results have been achieved in the past decade.
From a computational point of view, protein structure prediction and antigen interactions thereof is considered one of the main challenges in protein bioinformatics. Moreover, predictions become increasingly more challenging in the absence of robust and manually curated databases.
For this reason, to create robust and reliable protein prediction algorithms, it’s also crucial that researchers keep investing in the elucidation of the biochemical properties and sequences of new antibodies.
This extensive characterization can be performed using mass spectrometry, X-ray crystallography, and next-generation sequencing technologies. Nevertheless, producing crystalline antibodies is considered challenging to the complexity of its structure and its conformational heterogeneity.
However, technical progress has created the opportunity to elucidate the structure of multiple antibodies. Moreover, the creation of dedicated and curated antibody-specific databases continues to be the most important source of data for immunoinformatics.
One of the most important databases available today is the international ImMunoGeneTics information system (IMGT®). Created in 1989 by Marie-Paule Lefranc (CNRS, University of Montpellier, France), the IMGT® contains the genetic information of 7,235 antibodies and structural information for 6,621.
Concluding remarks
Antibody design is a particularly important stage of the antibody development pipeline. This stage aims to increase an antibody’s specificity, affinity, and stability, among others.
This optimization is recurrently achieved by using computational methods, directed evolution methods, or a combination of the two.
To date, the most successful strategies for antibody design rely on the combination of computational and directed evolution methodologies. Nevertheless, in recent years we have seen an increased interest in the development of robust de novo antibody design methods.
This interest continues to rise as more and more structural information becomes available and researchers strive to decrease animal use, costs and lengthy timeframes commonly associated with conventional antibody generation techniques.
- Harris, L. J. et al. Refined structure of an intact IgG2a monoclonal antibody. Biochemistry. 1997; 36(7):1581-1597. doi: 10.1021/bi962514+
- Lefranc, M.-P. Immunoglobulin (IG) and T cell receptor genes (TR): IMGT® and the birth and rise of immunoinformatics. Front Immunol. 2014; 5:22. doi: 10.3389/fimmu.2014.00022
- Rose, A. S. et al. NGL viewer: web-based molecular graphics for large complexes. Bioinformatics doi:10.1093/bioinformatics/bty419
- Sifniotis, V. et al. Current Advancements in Addressing Key Challenges of Therapeutic Antibody Design, Manufacture, and Formulation. Antibodies. 2019; 8:36. doi: 10.3390/antib8020036
- Stoevesandt, O. and Taussig, M. J. Phospho-specific antibodies by design. Nat Biotechnol. 2013; 31(10):889-891. doi: 10.1038/nbt.2712
- Tiller, K. E. and Tessier, P. M. Advances in Antibody Design. Annu Rev Biomed Eng. 2015; 17: 191–216. doi: 10.1146/annurev-bioeng-071114-040733
- Yamashita, T. Toward rational antibody design: recent advancements in molecular dynamics simulations. Int Immunol. 2018; 30(4): 133–140, doi: 10.1093/intimm/dxx077