 Antibody production
Antibody production
COVID-19 antibodies, antigens, and services
SARS-CoV-2 is a new Betacoronavirus responsible for the COVID-19 respiratory outbreak. The virus is also the third of the genus known to have been transmitted from animals to humans and to develop a sustained human-to-human transmission. The outbreak, first reported in Wuhan (China) on December 31, 2019, has now infected more than 1.2 million around the world and caused the deaths of more than 70 000 people (data from April 7, 2020).
These devastating numbers continue to push the scientific community in the search for therapeutic, diagnostic, and prophylactic solutions. But as most countries continue to impose lock downs and travel restrictions due to the COVID-19 pandemic, many research institutions have been forced to put their activities on hold.
Services dedicated to support COVID-19 research
To mitigate the impact of the new pandemic, it is important to develop rapid point-of-care tests and treatment options for severe cases of the disease. This will alleviate the burden on the health care system in the short-term and give us time to develop long-term vital strategies such as vaccination.
Our team is now focused on supporting researchers on the development of antibodies, prophylactic vaccines, and immunoassays for research and diagnostics.
Antibody Development
- Accelerated therapeutic antibody discovery by screening a naïve antibody human library (LiAb-SFMAX™, scFv, Fab, IgG, >5×1010) or an immune human antibody library (obtained from the plasma of COVID-19 survivors) using phage display
- Accelerated analytical antibody discovery by screening a naïve high diversity rabbit library (LiAb-SFRab™, scFv, Fab, IgG, >1×1010) using phage display
Associated services: Phage display
Vaccine development
- Accelerated vaccine construct design and production using our fast peptide synthesis service and high-throughput protein expression screening system
Associated services: Peptide synthesis and Protein expression
Immunoassay development
- Accelerated sandwich ELISA development using our ready-made anti-SARS-CoV-2 antibodies for research and diagnostics
- Custom sandwich ELISA development using our fast-track solutions to discover new antiviral antibodies
Associated services: Sandwich ELISA development
Custom gene synthesis
- Fast-track synthesis of gene, primers, and plasmid synthesis – optimized vector design
Associated services: Gene synthesis
Off-the-shelf stocks of SARS-CoV-2 antigens and antibodies
The first genomic sequence of the new pathogenic virus was published on January 24, providing essential data for the development of targeted therapies, prophylactic treatments, and diagnostics solutions. At ProteoGenix, we are making use of this data to produce essential custom antigens and antibodies to support COVID-19 related research.
Structural protein antigens
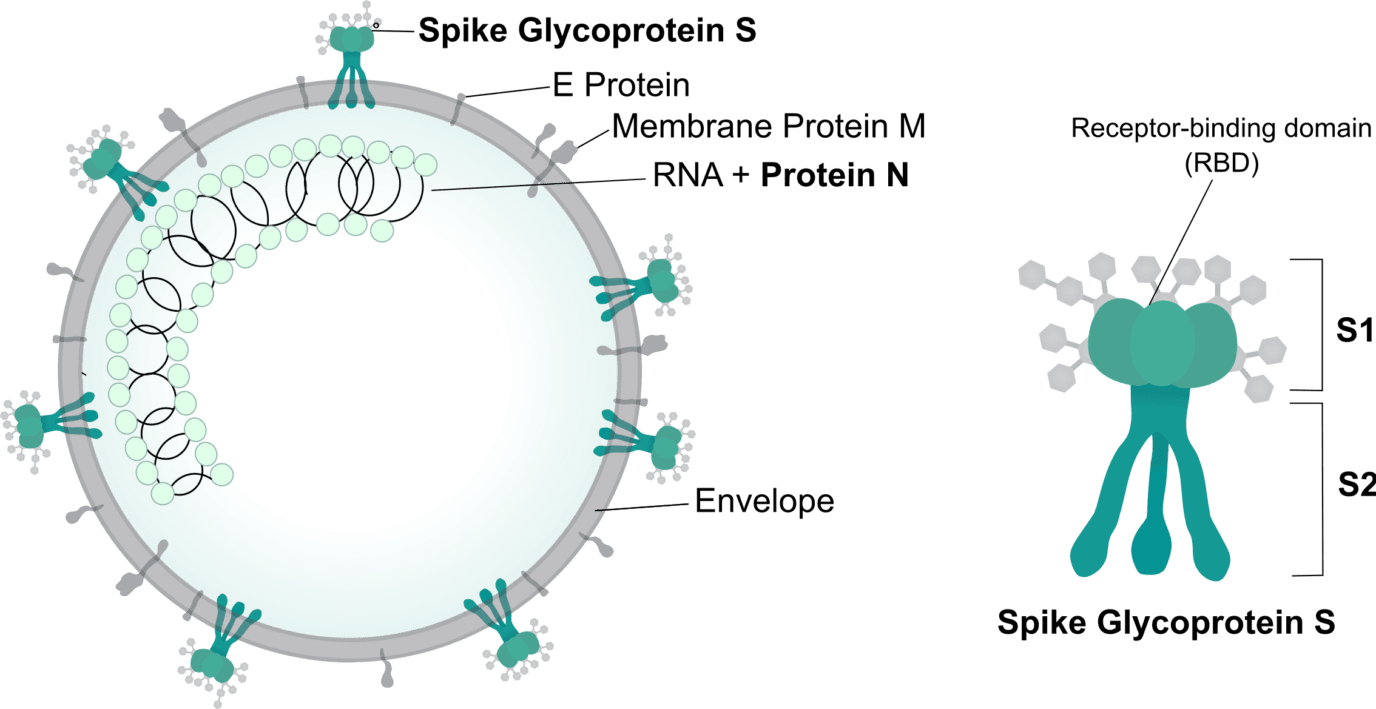
The structural proteins of SARS-CoV-2 are well characterized and considered attractive targets for new therapies. One of the leading targets of current drug discovery efforts is the spike glycoprotein S. As observed in other strains of coronavirus, the spike protein is responsible for binding the human receptor ACE2 and mediates the virus’ internalization. The protein consists of a S1 and S2 subunit. The S1 contains the receptor-binding domain (RBD) which interacts directly with ACE2, while the S2 induces the fusion of the viral envelop with the cellular membrane. Both subunits and the RBD are considered attractive targets for therapeutic development.
The nucleoprotein (protein N) is one of the most abundant proteins in coronavirus and deemed essential for its efficient replication. For this reason, protein N is considered a good target for the development of targeted diagnostic tools. Moreover, drugs designed to target this protein may interfere with SARS-CoV-2 replication and thus reduce the impacts of a widespread infection.
Non-structural protein antigens
Some non-structural proteins encoded in the genome of SARS-CoV-2 (GenBank accession number MN908947) may constitute important additional targets for research, therapy, and diagnostics. Their suitability as targets has been demonstrated in studies with other coronavirus strains. Stocks of these proteins or antigenic fragments thereof are available for ordering:
| Structure | Name | Role |
|---|---|---|
 |
3C-like proteinase (3CLp) | It is the main proteinase in coronaviruses, crucial for processing viral proteins |
 |
Papain-like proteinase (nsp3) | It is a vital multifunctional protein responsible for processing viral proteins |
 |
Growth factor-like peptide (nsp10) | Plays an essential role in viral replication |
 |
2’-O-methyltransferase (nsp16) | It is responsible for performing the methylation of viral mRNA, an essential viral mechanism for evading the host’s immune system |
 |
RNA-directed RNA polymerase (RdRp) | Vital for the viral replication cycle |
Concluding remarks
As an active member of the research community, ProteoGenix has anticipated the needs of researchers working on COVID-19. Our continuous efforts to develop and provide the most relevant antigens and antibodies are aimed at accelerating essential research against this new pathogenic strain.
We will remain mobilized by your side to ensure that all your COVID-19 projects stay on track.
- Almazán, F. et al. The nucleoprotein is required for efficient coronavirus genome replication. J Virol. 2004; 78(22):12683-12688. doi: 10.1128/JVI.78.22.12683-12688.2004
- Andersen, K. G. et al. The proximal origin of SARS-CoV-2. Nat Med. 2020. doi: 10.1038/s41591-020-0820-9
- Angeletti, S. et al. COVID‐2019: The role of the nsp2 and nsp3 in its pathogenesis. J Med Virol. 2020. doi: 10.1002/jmv.25719
- Lei, J. et al. Nsp3 of coronaviruses: Structures and functions of a large multi-domain protein. Antiviral Res. 2018; 149:58-74. doi: 10.1016/j.antiviral.2017.11.001
- Zhang, C. et al. Protein structure and sequence re-analysis of 2019-nCoV genome refutes snakes as its intermediate host and the unique similarity between its spike protein insertions and HIV-1. J Proteome Res. 2020. doi: 10.1021/acs.jproteome.0c00129
- Zhang, L. et al. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020; pii: eabb3405. doi: 10.1126/science.abb3405
- Zhavoronkov, A. et al. Potential COVID-2019 3C-like Protease Inhibitors Designed Using Generative Deep Learning Approaches. ChemRxiv. Preprint. 2020. doi: 10.26434/chemrxiv.11829102.v2